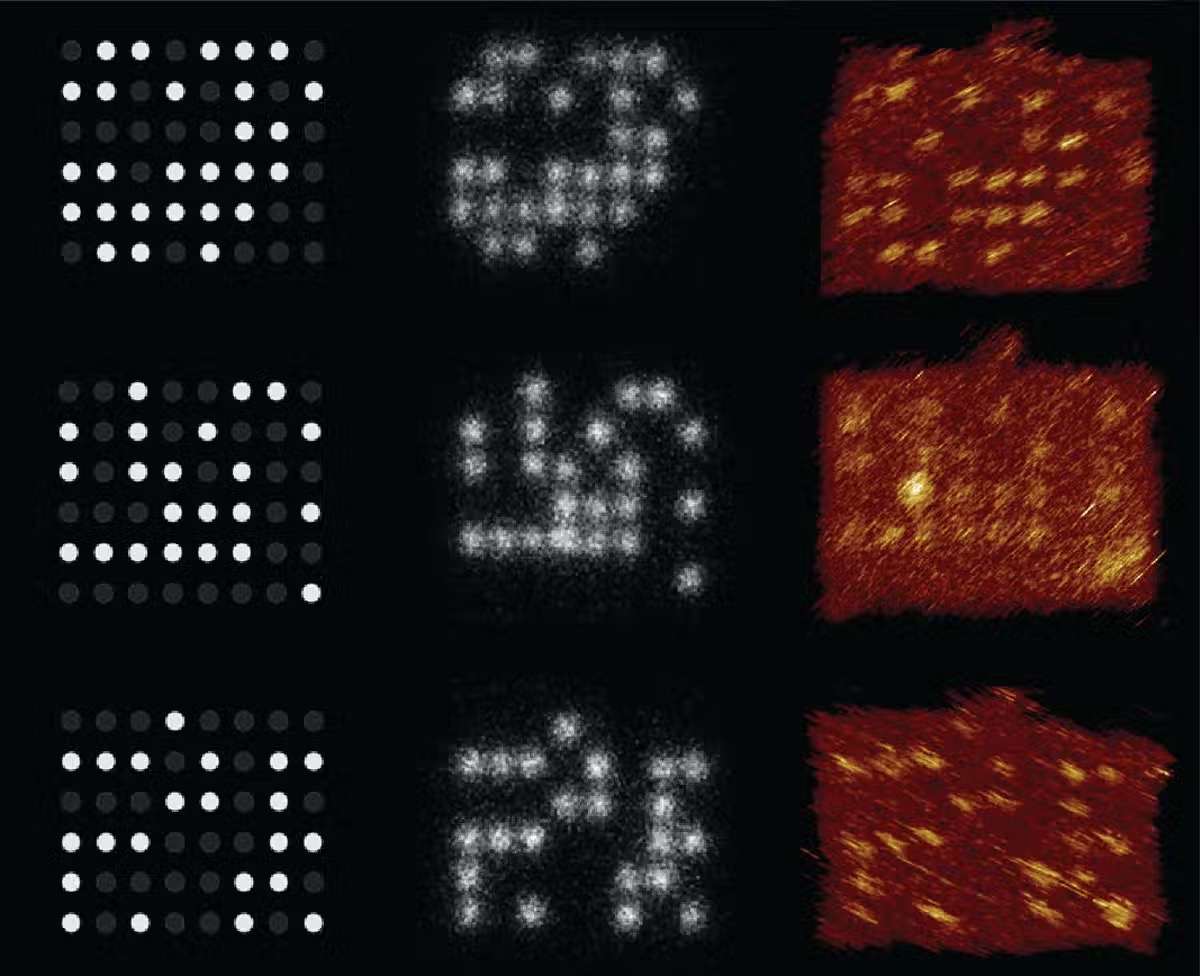
We and our partners have fostered a method for putting away information utilizing stakes and pegboards made from DNA and recovering the information with a magnifying instrument – a sub-atomic variant of the Lite-Brite toy. Our model stores data in designs utilizing DNA strands separated around 10 nanometers separated. Ten nanometers is in excess of multiple times less than the breadth of a human hair and multiple times less than the measurement of a bacterium.
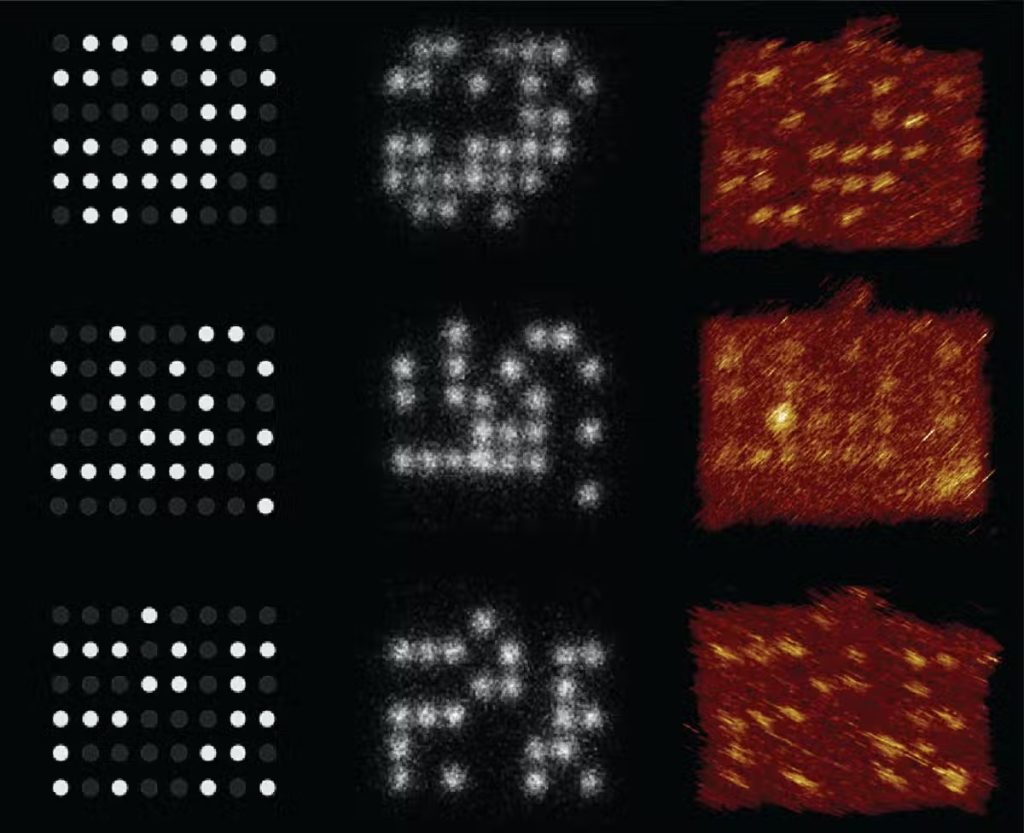
We tried our computerized nucleic corrosive memory (dNAM) by putting away the proclamation “Information is in our DNA!\n.” We portrayed the examination in a paper distributed in the diary Nature Communications on April 22, 2021.
Past strategies for recovering information in DNA require the DNA to be sequenced. Sequencing is the method involved with perusing the hereditary code of strands of DNA. However it is a useful asset in medication and science, it wasn’t planned considering DNA memory.
Our methodology utilizes a magnifying lens to optically peruse the information. Since the DNA stakes are situated nearer than a portion of the frequency of noticeable light, we utilized super-goal microscopy, which evades the diffraction furthest reaches of light. This gives a method for perusing the encoded information without sequencing the DNA.
The examples of DNA strands – the stakes – light up when fluorescently named DNA tie to them. Since the fluorescent strands are short, they quickly tie and unbind. This makes them squint, making it simpler to isolate one stake from another and read the put away data. We utilize the fluorescent examples of every pegboard as a code to store lumps of information.
The magnifying lens can picture countless the DNA stakes in a solitary recording, and our mistake remedy calculations guarantee we recuperate the entirety of the information. Subsequent to representing the pieces utilized by the calculations, our model had the option to peruse information at a thickness of 330 gigabits for each square centimeter.
Why it is important
You’re not prone to have a DNA stockpiling gadget in your telephone or PC, essentially at any point in the near future. DNA information capacity is promising for chronicled capacity – putting away a lot of data for extensive stretches of time. DNA can store a ton of data in a little space. It would be feasible to store each tweet, email, photograph, tune, film and book at any point made in a volume comparable to a gems box. Furthermore, information put away in DNA could keep going for a really long time, considering that the biomolecule has a half-existence of north of 500 years.
What other exploration is being finished
Scientists have been creating strategies for putting away information in DNA for quite some time. Those strategies include the plan and blend of remarkable strings of data produced using the DNA nucleotides adenine (A), thymine (T), cytosine (C) and guanine (G). This data is recuperated by perusing the strings utilizing sequencing innovation.
What’s straightaway
From here, we want to build how much information that we can store in dNAM, decline how much time it takes to compose and peruse the information, and utilize the method to encode information.