Since researchers previously planned the total human genome, consideration has now gone to the subject of how cells utilize this expert duplicate of hereditary guidelines. It is known that when qualities are turned on, portions of the DNA arrangements in the phone core are replicated into more limited string-like atoms, RNA, that convey the particles fundamental for endurance and cell-explicit capabilities.
Understanding the profiles of RNA in a cell can show which qualities are dynamic and permit scientists to conjecture what the cell is doing. The innovation for estimating RNA by enormously equal DNA sequencer, RNA-sequencing, has turned into a standard procedure over the course of the last 10 years. All the more as of late, fast mechanical advances license RNA sequencing at the single-cell level from huge number of cells in equal, speeding up progress in the biomedical sciences. In any case, measuring RNAs from such a small material stances extraordinary specialized difficulties. Indeed, even with cutting edge hardware, information created from single-cell RNA sequencing information contain huge location mistakes, including the supposed “drop-out impact.” Moreover, even little blunders in the estimations for an enormous number of qualities can rapidly add up so any helpful data is lost among signal clamor.
Presently, a group from the Kyoto University Institute for Advanced Study of Human Biology (WPI-ASHBi) has fostered another numerical technique that can kill the commotion and in this manner empower the extraction of clear signals from single-cell RNA sequencing information. The new technique effectively diminishes irregular examining clamor in the information to empower an exact and finish comprehension of a cell’s action. The exploration has as of late been distributed in the diary Life Science Alliance.
The lead creator of the paper, Yusuke Imoto from ASHBi, makes sense of, “Every quality addresses an alternate aspect in RNA sequencing information, and that implies that huge number of aspects should be gathered across various cells and dissected. Indeed, even the smallest commotion in one aspect can significantly affect the downstream information examinations with the goal that possibly significant signs are lost. To this end we call this the ‘scourge of dimensionality.'”
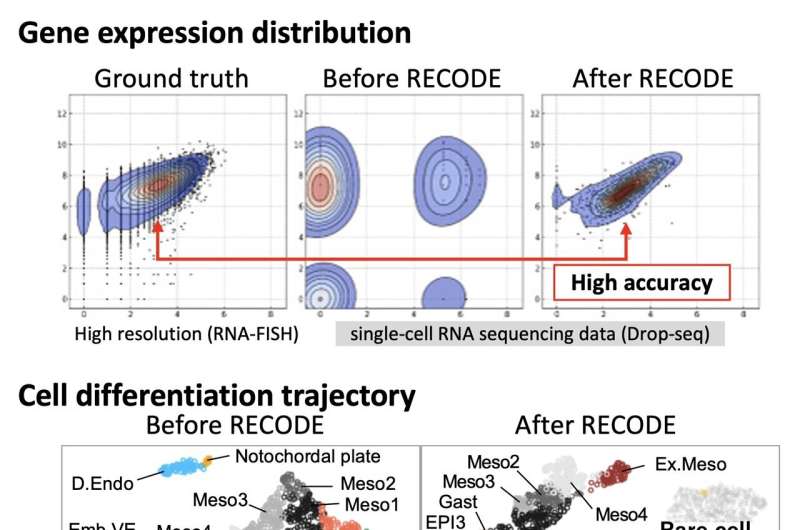
To break the scourge of dimensionality, the Kyoto group has fostered another sound decrease technique, RECODE — meaning “goal of the scourge of dimensionality” — to eliminate the arbitrary testing commotion from single-cell RNA sequencing information. RECODE applies high-layered factual hypotheses to recuperate exact outcomes, in any event, for qualities communicated at exceptionally low levels.
In the first place, the group tried their strategy on information from an extensively very much concentrated on cell populace, human fringe blood. They affirmed that RECODE effectively eliminates the scourge of dimensionality to uncover articulation designs for individual qualities near their normal qualities.
Then, when looked at against other cutting edge examination techniques, RECODE outflanked the opposition by giving a lot more genuine portrayals of quality enactment. Besides, RECODE is less complex to use than different techniques, without depending on boundaries or utilizing AI for the computations to work.
At long last, the group tried RECODE on a complex dataset from mouse undeveloped organism cells containing various sorts of cells with exceptional quality articulation designs. While different strategies obscured the outcomes, RECODE obviously settled quality articulation levels, in any event, for uncommon cell types.
According to imoto, “Single-cell RNA sequencing information investigation remains in fact testing and is a creating procedure, however our RECODE calculation is a stage towards having the option to uncover the genuine ways of behaving of single-cell structures. With our commitment, single-cell RNA sequencing information examination could turn into a strong exploration instrument with enormous ramifications across numerous organic fields.”
One more driving creator Tomonori Nakamura, a scientist from ASHBi and The Hakubi Center for Advanced Study, Kyoto University, adds, “By opening the genuine force of single-cell RNA sequencing, RECODE will empower specialists to find unidentified uncommon cell types, prompting the turn of events and foundation of the new examination field in fundamental science as well as clinical application and medication revelation research.”