At the point when the Human Genome Project reported that they had finished the principal human genome in 2003, it was a pivotal achievement – interestingly, the DNA outline of human existence was opened. In any case, it accompanied a catch – they weren’t really ready to assemble all the hereditary data in the genome. There were holes: unfilled, frequently tedious areas that were excessively confounding to sort out.
With progressions in innovation that could deal with these redundant successions, researchers at last filled those holes in May 2021, and the main start to finish human genome was formally distributed on Mar. 31, 2022.
I’m a genome scholar who concentrates on dull DNA groupings and how they shape genomes all through developmental history. I was essential for the group that described the recurrent successions missing from the genome. What’s more, presently, with a genuinely complete human genome, these uncovered dull districts are at last being investigated in full interestingly.
The lacking riddle parts
German botanist Hans Winkler begat “genome” in 1920, consolidating “quality” with the postfix “- ome,” signifying “complete set,” to portray the full DNA arrangement held inside every cell. Specialists actually utilize this word a century after the fact to allude to the hereditary material that makes up a creature.
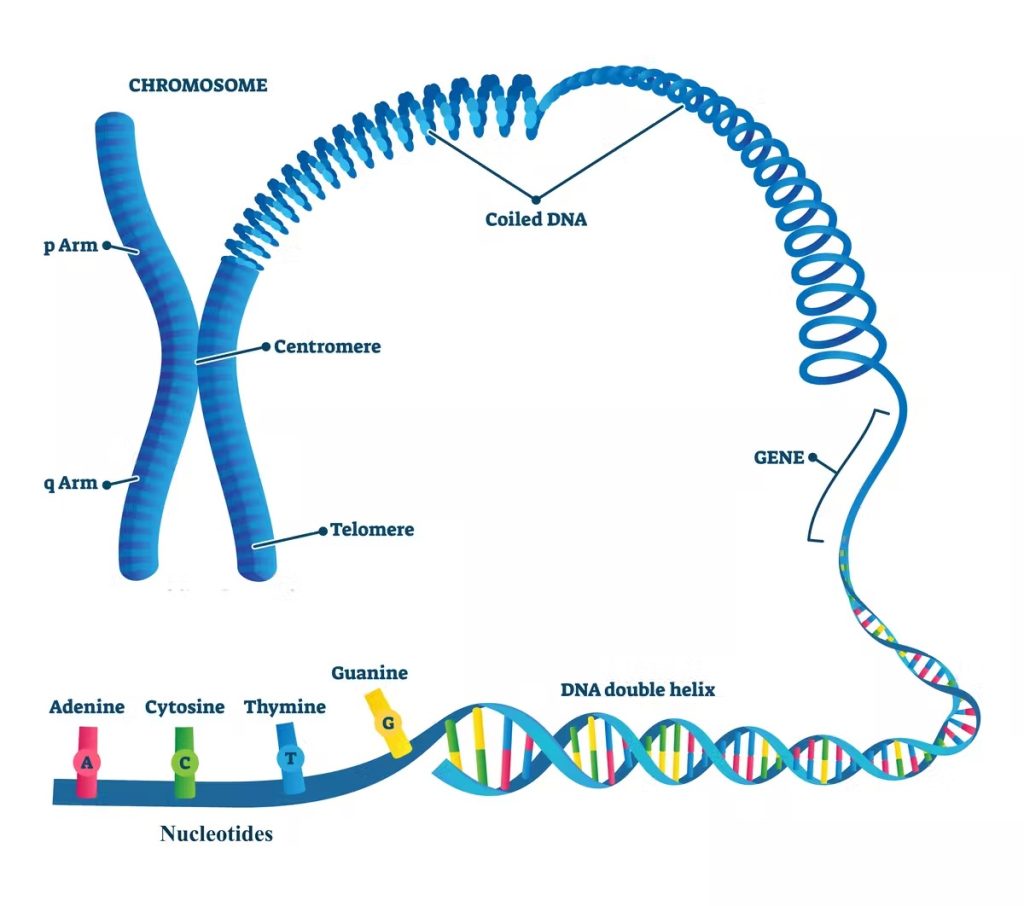
One method for portraying what a genome resembles is to contrast it with a reference book. In this relationship, a genome is a treasury containing the DNA directions forever. It’s made out of a huge swath of nucleotides (letters) that are bundled into chromosomes (parts). Every chromosome contains qualities (passages) that are districts of DNA which code for the particular proteins that permit a living being to work.
While each living life form has a genome, the size of that genome differs from species to species. An elephant involves a similar type of hereditary data as the grass it eats and the microscopic organisms in its stomach. However, no two genomes look precisely similar. Some are short, similar to the genome of the bug staying microbes Nasuia deltocephalinicola with only 137 qualities across 112,000 nucleotides. Some, similar to the 149 billion nucleotides of the blooming plant Paris japonica, are long to the point that it’s hard to get a feeling of the number of qualities that are held inside.
Yet, qualities as they’ve customarily been perceived – as stretches of DNA that code for proteins – are only a little piece of an organic entity’s genome. They make up under 2% of human DNA, as a matter of fact.
The human genome contains approximately 3 billion nucleotides and just shy of 20,000 protein-coding qualities – an expected 1% of the genome’s absolute length. The leftover almost 100% is non-coding DNA groupings that don’t create proteins. Some are administrative parts that work as a switchboard to control how different qualities work. Others are pseudogenes, or genomic relics that have lost their capacity to work.
Also, over portion of the human genome is dull, with different duplicates of close indistinguishable groupings.
What is monotonous DNA?
The easiest type of tedious DNA are blocks of DNA rehashed again and again couple called satellites. While how much satellite DNA a given genome has fluctuates from one individual to another, they frequently bunch toward the finishes of chromosomes in locales called telomeres. These locales safeguard chromosomes from debasing during DNA replication. They’re likewise found in the centromeres of chromosomes, a locale that helps watch out for hereditary data when cells partition.
Specialists actually come up short on clear comprehension of the relative multitude of elements of satellite DNA. But since satellite DNA structures one of a kind examples in every individual, measurable scientists and genealogists utilize this genomic “finger impression” to match crime location tests and track lineage. More than 50 hereditary issues are connected to varieties in satellite DNA, including Huntington’s illness.
One more plentiful sort of monotonous DNA are transposable components, or arrangements that can move around the genome.
A few researchers have depicted them as childish DNA since they can embed themselves anyplace in the genome, no matter what the outcomes. As the human genome advanced, numerous transposable groupings gathered changes quelling their capacity to move to keep away from hurtful interferences. In any case, some can almost certainly still move about. For instance, transposable component inclusions are connected to various instances of hemophilia A, a hereditary draining problem.
In any case, transposable components aren’t simply problematic. They can have administrative capabilities that assist with controlling the declaration of other DNA arrangements. At the point when they’re amassed in centromeres, they may likewise assist with keeping up with the respectability of the qualities crucial to cell endurance.
They can likewise add to advancement. Scientists as of late found that the inclusion of a transposable component into a quality critical to improvement may be the reason a few primates, including people, never again have tails. Chromosome improvements because of transposable components are even connected to the beginning of new species like the gibbons of southeast Asia and the wallabies of Australia.
Finishing the genomic puzzle
As of not long ago, a significant number of these perplexing districts might measure up to the furthest side of the moon: known to exist, however inconspicuous.
At the point when the Human Genome Project previously sent off in 1990, mechanical constraints made it difficult to reveal monotonous districts in the genome completely. Accessible sequencing innovation could learn about 500 nucleotides all at once, and these short sections needed to cover each other to reproduce the full grouping. Scientists utilized these covering sections to recognize the following nucleotides in the grouping, gradually expanding the genome gathering each part in turn.
These monotonous hole districts resembled assembling a 1,000-piece puzzle of a cloudy sky: When each piece appears to be identical, how do you have any idea where one cloud begins and another closures? With close indistinguishable covering extends in many spots, completely sequencing the genome by piecemeal became impossible. A large number of nucleotides stayed secret in the primary emphasis of the human genome.
From that point forward, grouping patches have slowly filled in holes of the human genome little by little. What’s more, in 2021, the Telomere-to-Telomere (T2T) Consortium, a worldwide consortium of researchers attempting to finish a human genome gathering from one finish to another, declared that all excess holes were at last filled.
This was made conceivable by improved sequencing innovation fit for perusing longer groupings great many nucleotides long. With more data to arrange dreary successions inside a bigger picture, it became simpler to distinguish their legitimate spot in the genome. Like improving on a 1,000-piece puzzle to a 100-piece puzzle, long-read successions made it conceivable to collect huge monotonous districts interestingly.
With the rising force of long-perused DNA sequencing innovation, geneticists are situated to investigate another period of genomics, unwinding complex dreary arrangements across populaces and species interestingly. Furthermore, a total, hole free human genome gives a significant asset to specialists to research monotonous districts that shape hereditary construction and variety, species development and human wellbeing.
Yet, one complete genome doesn’t catch everything. Endeavors keep on making different genomic references that completely address the human populace and life on Earth. With more complete, “telomere-to-telomere” genome references’, how researchers might interpret the redundant dim matter of DNA will turn out to be all the more clear.